2.3. Importing data into DataFrames and exploring its attributes#
pandas provides many functions to import data into dataframes, such as read_csv() to read delimited text files, or read_excel() for Excel or OpenDocument spreadsheets. read_csv() provides options that allow you to filter the data, such as specifying the separator/delimiter, the lines that form the headers, which rows to skip, etc. Let’s analyze the mineral_properties.txt. Below a screenshot of it:
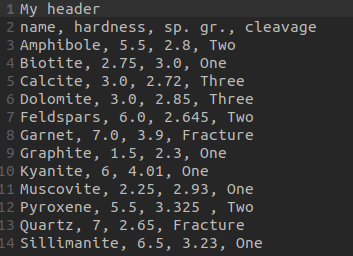
below we import the .txt:
we indicate that the separator is the comma
"sep=','"we indicate the header (what should be the columns names) is in the second line
"header=[1]"we indicate to not skip any rows
"skiprows=None"we indicate the first column should be the index of the rows
"index_col=0"
file_location = ("mineral_properties.txt")
df4 = pd.read_csv(file_location, sep=',', header=[1],
skiprows=None, index_col=0)
df4
| hardness | sp. gr. | cleavage | |
|---|---|---|---|
| name | |||
| Amphibole | 5.50 | 2.800 | Two |
| Biotite | 2.75 | 3.000 | One |
| Calcite | 3.00 | 2.720 | Three |
| Dolomite | 3.00 | 2.850 | Three |
| Feldspars | 6.00 | 2.645 | Two |
| Garnet | 7.00 | 3.900 | Fracture |
| Graphite | 1.50 | 2.300 | One |
| Kyanite | 6.00 | 4.010 | One |
| Muscovite | 2.25 | 2.930 | One |
| Pyroxene | 5.50 | 3.325 | Two |
| Quartz | 7.00 | 2.650 | Fracture |
| Sillimanite | 6.50 | 3.230 | One |
Note that if we try to call any of the columns from df4 we will get an error.
df4['hardness']
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File c:\Users\gui-win10\AppData\Local\pypoetry\Cache\virtualenvs\learn-python-rwbttIgo-py3.11\Lib\site-packages\pandas\core\indexes\base.py:3805, in Index.get_loc(self, key)
3804 try:
-> 3805 return self._engine.get_loc(casted_key)
3806 except KeyError as err:
File index.pyx:167, in pandas._libs.index.IndexEngine.get_loc()
File index.pyx:196, in pandas._libs.index.IndexEngine.get_loc()
File pandas\\_libs\\hashtable_class_helper.pxi:7081, in pandas._libs.hashtable.PyObjectHashTable.get_item()
File pandas\\_libs\\hashtable_class_helper.pxi:7089, in pandas._libs.hashtable.PyObjectHashTable.get_item()
KeyError: 'hardness'
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In[67], line 1
----> 1 df4['hardness']
File c:\Users\gui-win10\AppData\Local\pypoetry\Cache\virtualenvs\learn-python-rwbttIgo-py3.11\Lib\site-packages\pandas\core\frame.py:4090, in DataFrame.__getitem__(self, key)
4088 if self.columns.nlevels > 1:
4089 return self._getitem_multilevel(key)
-> 4090 indexer = self.columns.get_loc(key)
4091 if is_integer(indexer):
4092 indexer = [indexer]
File c:\Users\gui-win10\AppData\Local\pypoetry\Cache\virtualenvs\learn-python-rwbttIgo-py3.11\Lib\site-packages\pandas\core\indexes\base.py:3812, in Index.get_loc(self, key)
3807 if isinstance(casted_key, slice) or (
3808 isinstance(casted_key, abc.Iterable)
3809 and any(isinstance(x, slice) for x in casted_key)
3810 ):
3811 raise InvalidIndexError(key)
-> 3812 raise KeyError(key) from err
3813 except TypeError:
3814 # If we have a listlike key, _check_indexing_error will raise
3815 # InvalidIndexError. Otherwise we fall through and re-raise
3816 # the TypeError.
3817 self._check_indexing_error(key)
KeyError: 'hardness'
Do you know why?
Answer …
In case you were not able to answer the above question, let’s look into df4.columns
df4.columns
Index([' hardness', ' sp. gr.', ' cleavage'], dtype='object')
You see there are spaces at the beginning of each column name… this happens because that’s how people usually type, with commas followed by a space. We could use the skipinitialspace = True from the pd.read_csv() function to avoid this. Let’s try it out:
df4 = pd.read_csv(file_location + 'mineral_properties.txt',sep=',',header=[1],
skiprows=None, index_col=0, skipinitialspace=True)
print(df4.columns)
Index(['hardness', 'sp. gr.', 'cleavage'], dtype='object')
Ok, much better!
